Basics
CoPolDB is a database that focuses on radical polymerization and manages information on the monomer pairs that form copolymers, as well as their reactivity ratios obtained from literature. Additionally, CoPolDB plans to handle information through machine learning simulations and data registration from researchers in the future. The database's main goal is to provide an integrated and easily accessible source of data on copolymerization, where monomer, copolymer, and reactivity ratio information are linked to each other.
Currently, CoPolDB consists of two parts, Monomers and Copolymers, which can be accessed from the top menu.
- The "Monomers" part provides information on the monomers used to form copolymers, including identifiers such as SMILES (Simplified Molecular Input Line Entry System) and InChI (International Chemical Identifier), monomer names. Each monomer entry is linked to the corresponding copolymer and copolymerization entries. If a monomer is registered in PubChem, the monomer name and identifier information are maintained to match the PubChem information. In addition to the name and identifiers, various other information such as molecular formula, molecular weight and structural formula calculated by RDKit are displayed.
- The "Copolymers" part provides information on copolymers and monomer reactivity ratios (r1 and r2). The features of this part are (1) reactivity ratios obtained from different literatures for a copolymer are integrated on a single page, providing the function to display scatter plots and F1 / f1 plots for comparison, and (2) DOI (Digital Object Identifiers) links are provided to copolymerization references for easy access to experimental information.
Search
CoPolDB offers two types of search functions to enable easy access to desired information. The first is a simple search function that searches by monomer name, which is accessible from the search box located at the top right of the "Browse" pages for both the "Monomers" and "Copolymers" parts. The search is based on conventional or IUPAC names of the monomers. The second search function is a detailed search that allows searching with multiple criteria, such as molecular formula and SMILES, and can be accessed from the "Search" page. The search function is available on the four pages listed below.
- Monomers' Browse page for search by name (#)
- Monomers' Search page for search by multiple conditions (#)
- Copolymers' Browse page for search by name (#)
- Copolymers' Search page for search by multiple conditions (#)
Pagination
On the "Browse" pages, a pagination bar appears above and below the list if the number of displayed items exceeds 20. The pagination bar consists of a Previous button, a Next button, and buttons for up to five page numbers, where the page button of the current page is highlighted in blue. The pagination provides fast forward and fast rewind functions.
- To fast-forward through the pages, double-click the "Next" button.
- To fast-rewind through the pages, double-click the "Previous" button.
The current page number and the total number of pages are displayed in the format "{current page} / {total number of pages}" at the top center of the list. The same is displayed at the bottom of the list.
Sort
The list displayed on the "Browse" pages can be sorted using the two drop-downs order and by in the upper left corner.
- The "by" drop-down allows you to select a display item (e.g., monomer name, molecular weight) in the list entries.
- The "order" drop-down is used to specify the order in which the listing is displayed, which is common to both the Monomers' and Copolymers' "Browse" pages and has the three items: default, ascending and descending. When "default" is selected, the list is ordered by CoPolDB's internal ID.
Monomers
As mentioned above, the "Monomers" part consists of four tabbed pages: Overview, Browse, Search and Statistics. The "Monomers' Overview" page provides a chemical description of monomer, the "Monomers' Browse" shows a list of monomers, the "Monomers' Search" page provides interfaces for inputting search criteria, and the "Monomers' Statistics" provides basic statistical information. To switch the pages, click on the tab above the "Monomers" part.
Browse
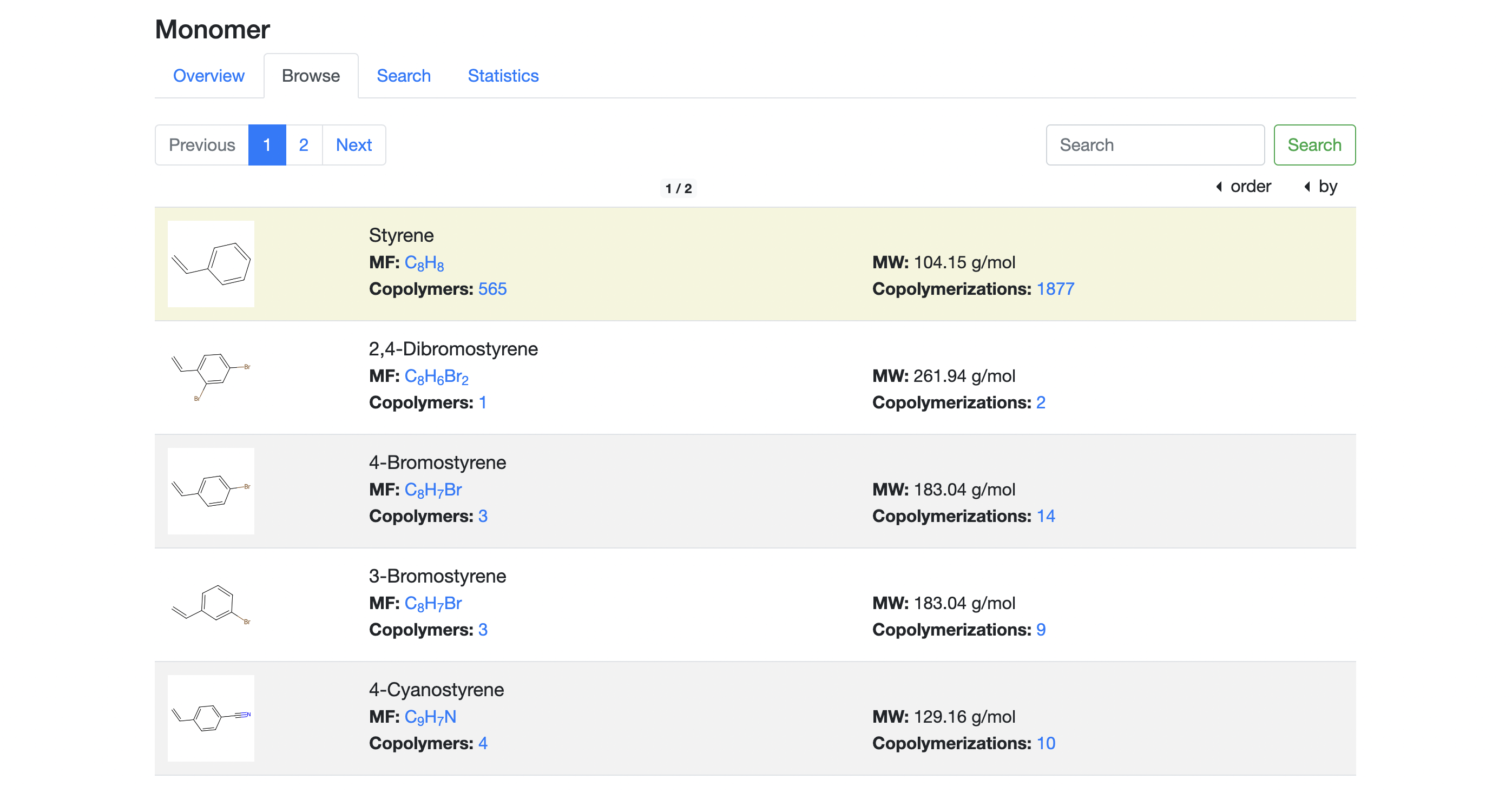
The "Monomers' Browse" page provides the functions of searching for monomers by name sorting and pagination. To cancel search, sorting, or pagination, click the "Browse" tab on the top.
- Search: enter a conventional name or IUPAC name of monomer in the search box in the upper left corner of the entry list and click the "Search" button to search for the monomer of interest. If you cannot find the monomer, try a search by other conditions such as molecular formula on the "Search" page.
- Sorting: select "ascending" or "descending" in the "order" drop-down and then select the item you wish to sort by in the "by" drop-down in the upper right corner of the entry list. The list can be sorted by monomer name, molecular formula, molecular weight, number of copolymers or number of copolymerizations.
- Pagination: click a page number, the "Previous" button or the "Next" button to show the monomer list for the page. By double-clicking on the "Next" button, moving forward five pages is performed. Likewise, by double-clicking the "previous" button, moving back five pages is performed.
In the page, each list entry displays the following information:
- 2-dimensional structural formula: a PNG image generated by RDKit based on SMILES is shown. Click the image to open the detailed information of the monomer.
- Monomer name: a conventional name of the monomer is displayed. If the conventional name is unknown, an IUPAC name will be displayed. If the SMILES of the monomer is registered in PubChem, then the conventional name managed by PubChem is displayed.
- Molecular formula: the molecular formula generated by RDKit based on the SMILES is displayed. Clicking on the formula will display the "Browse" page listing monomers with the same formula.
- Molecular weight: the molecular weight computed by RDKit based on the SMILES is displayed.
- The number of copolymers using the monomer: the number of copolymers linked to the monomer is displayed. Clicking on the number will open the "Browse" page of the corresponding copolymers.
- The number of copolymerizations using the monomer: the number of copolymerizations linked to the monomer is displayed. Clicking on the number will open the "Browse" page of the corresponding copolymerizations.
Detail Page
CoPolDB provides detailed pages for each monomer entry. To access a detailed page, click on the image of 2D structural formula shown to the left of an entry. The detailed page consists of five sections: (1) Identifiers, (2) Structures, (3) Molecular Formula and Computed Descriptors, (4) MOL File and (5) Similar Monomers.(1) Identifiers
In addition to the IUPAC name, SMILES (simplified molecular input line entry system) and InChI (IUPAC International Chemical Identifier) are shown for identifying the monomer. If the monomer is registered in PubChem, it is maintained to match the IUPAC name, canonical SMILES and isomeric SMILES in PubChem, while InChI and InCHI key are generated by Open Babel.(2) Structures
This section displays the 2D and 3D structures generated by RDKit and 3Dmol.js, respectively. To generate a conformation for the 3D structure, CoPolDB uses a distance geometry (DG) method called the Experimental-Torsion-Knowledge Distance Geometry (ETKDG). While the 2D structure is a normal PNG image, the 3D structure can be rotated by clicking and moving a mouse device and zoomed in and out by using the mouse wheel.(3) Molecular Formula and Computed Descriptors
This section shows the molecular formula and fifteen molecular descriptors calculated by RDKit, such as molecular weight, exact molecular weight, the number of valence electrons and the number of radical electrons.(4) MOL File
In this section, a MOL file is provided in text format. MOL is one of the de facto standards for data formats to describe in chemoinformatics. This MOL describes the results of the conformational calculation used to generate the 3D structures in the "Structures" section.(5) Similar Monomers
In this section, a list of monomers with similar structures will be displayed. Similarity is defined by Tanimoto similarity between Morgan fingerprints, which is one of the most popular fingerprints also known as circular fingerprints. Tanimoto similarity is defined as the ratio of the intersection of the two fingerprints over the union of them. In this section, monomers with similarity 0.25 or greater are displayed in descending order.Search
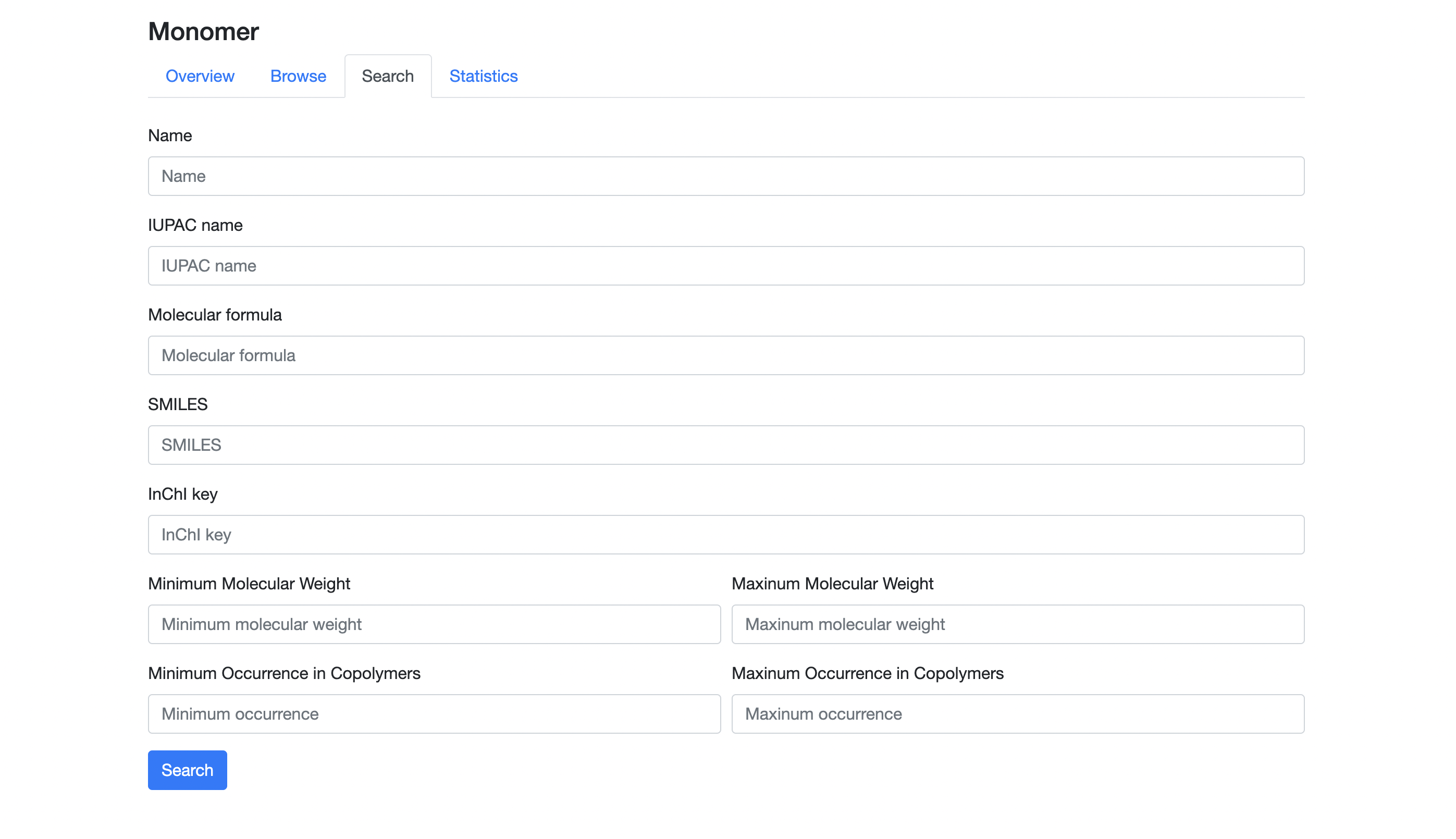
On the "Monomers' Search" page, you can search for monomers by specifying molecular formula or other conditions, while the browse page provides a search by monomer name only. This page provides an autocomplete feature. When you enter two or more characters in a text field for strings such as name, SMILES and molecular formula, up to 10 candidate strings will be displayed for you to choose one. If any candidate string does not exist in CoPolDB, the input field turns red and an error message is displayed. In this case, enter another string and confirm that the input field turns green.
- Name: enter a canonical or an IUPAC name in text.
- Molecular formula: enter a molecular formula in text, regardless of the sequence of atomic symbols. That is, the sequence of pairs of atomic symbols and their numbers is arbitrary; for example, C8H8 and H8C8 are treated as the same molecular formula.
- SMILES: enter a SMILES in text.
- InChI key: enter an InChI key in text.
- Minimum / Maximum molecular weight: enter the minimum and maximum values of molecular weights as integers.
- Minimum / Maximum occurrence in copolymers: enter the minimum and maximum values of the number of copolymerizations as integers.
Statistics
In the "Monomers' Statistics" page, 1) basic statistics and 2) histograms of monomers registered in CoPolDB are displayed.
- As basic statistics, the number of registered monomers and the number of monomers corresponding to PubChem records are displayed.
- As the histograms, the histogram of molecular weights and the histogram of occurrences in copolymers are displayed.
Copolymers
The "Copolymers" part is the core of CopolyDB and provides information on reactivity ratios for radical polymerization. The features are twofold: 1) the ability to compare the reactivity ratios of multiple copolymerizations obtained from one or more literatures for the same copolymer, and 2) if DOI (Digital Object Identifier) is assigned to a research paper, CoPolDB displays the link for easy access to the source information.
Throughout the "Copolymers" part of CoPolDB, the one monomer with the smaller molecular weight is referred to as the first monomer and the other monomer with the larger molecular weight is referred to as the second monomer. If the molecular weights are the same, the order is determined based on descriptor values other than molecular weight (HeavyAtomCount, NumValenceElectrons, etc.) calculated by RDKit.
This part, as well as the "Monomers" part, consists of four tabbed pages: Overview, Browse, Search and Statistics. The "Copolymers' Overview" page provides a chemical description of radical copolymerization, the "Copolymers' Browse" shows a list of copolymers and copolymerizations, the "Copolymers' Search" page provides input interfaces for obtaining specific copolymer list with multiple criteria, and the "Copolymers' Statistics" provides basic statistical information. To switch the pages, click on the tab above the "Monomers" part.
Browse
Unlike the "Monomers" part described above, there are two browse pages: Copolymers' Browse and Copolymerizations' Browse. The former page shows a list of monomer pairs with hyperlinks to one or more copolymerizations, while the latter page shows a list of reactivity ratios for individual copolymerization reactions. The two lists can be switched by clicking on the "Copolymerizations" or "Copolymers" button next to the tab menu. On either page, the first monomer is shown on the left side and the second monomer is shown on the right side in the entry.
Functions Common to Both Browse Pages
Both browse pages provide the functions of searching for copolymers or copolymerizations by name sorting and pagination. To cancel search, sorting, or pagination, click the "Browse" tab.- Search: enter a conventional or an IUPAC monomer name of one of two monomers in the search box in the upper left corner of the list, and then click the "Search" button to search for copolymers or copolymerizations. If the copolymer (or copolymerization) of interest is not found, try a search by other conditions such as molecular formula on the "Search" page.
- Sorting: select "ascending" or "descending" in the "order" drop-down and then select the item to be sorted in the "by" drop-down in the upper right corner of the list. The list can be sorted by monomer name, molecular formula, molecular weight, number of copolymers or number of copolymerizations.
- Pagination: click a page number, the "Previous" button or the "Next" button to show the monomer list for the page. By double-clicking on the "Next" button, you can move forward five pages. Likewise, by double-clicking the "previous" button, you can move back five pages.
Copolymers' Browse Page
In the page, each entry displays the following information:- Number of copolymerizations: the number of copolymerizations is displayed in the "Go into details" button on the left side. Click the button to open the detail page, where a list of reactivity ratios is displayed and you can also access literature via DOIs.
- 2-dimensional structural formulas: the structural formulas of the first and second monomer are displayed on the left and right, respectively. Click on the image of either structural formula to open a detailed page of the monomer.
- Monomer names: the names of the first and second monomer are displayed on the left and right sides, respectively. Click on either monomer name to open the detailed page of the monomer.
- Molecular formulas: the molecular formulas of the first and second monomer are displayed under the respective monomer name. Click on either formula to conduct a copolymer search by the formula.
- Molecular weights: the molecular weights of the first and second monomer are displayed under the respective molecular formula.
Copolymerizations' Browse Page
The browse page of copolymerizations displays a list of entries with the following items:- First monomer: The name of the first monomer is displayed. Click on the name to display the monomer detail page.
- Second monomer: The name of the second monomer is displayed. Click on the name to display the monomer detail page.
- r1: The reactivity ratio of the first monomer in the reference is displayed.
- r2: The reactivity ratio of the second monomer in the reference is displayed.
- Reference: The reference information and DOI link button are shown at the bottom of the entry. If the DOI is valid, the button is blue and clicking it will open the article page. If the DOI does not exist or is unknown, the button is gray and cannot be clicked.
Detail Page
The detailed page consists of two sections: (1) Copolymer Information, (2) Reactivity Ratios and (3) Alternative Monomers.(1) Copolymer Information
In this section, basic information about the two monomers that form a copolymer is presented in table format. The table consists of two columns, with first monomer information on the left and second monomer information on the right, and contains the following information.- Name: name for each monomer
- IUPAC name: IUPAC name for each monomer
- Molecular weight: molecular weight calculated by RDKit for each monomer
- Molecular formula: molecular formula generated by RDKit for each monomer
- 2D Structure: 2D structural formula generated by RDKit for each monomer
(2) Reactivity Ratios
All copolymerization reactions of the corresponding copolymer registered in CoPolDB can be displayed together in two plots and one reference table: an F1 / f1 plot, a scatter plot and a reference table consisting of monomer reactivity ratios and reference information. These three components can be synchronized in the way they are displayed by switching the "All" and "One" buttions in the upper left corner of the section. "All" can be used to plot all experimental data , and "One" can be used to select only one experiment of interest. To selectively display reactivity ratios, click on the number on the legend of the plots or on the table.- F1 / f1 plot: for each copolymerization reaction, a plot is displayed according to Equations (5) and (7) on the Overview page, with the ranges of both axes, f1 and F1, as [0,1].
- Scatter plot: a scatter plot is displayed with r1 on the horizontal axis and r2 on the vertical axis.
- Reference table: Reactivity ratios are displayed in the upper part of each row as well as 95% confidence limits, and references and DOIs in the lower part of each row. If a reactivity ratio or confidence limit is not available, "---" will be displayed. By clicking on the number button to the right of each row, the corresponding data of the F1 / f1 and scatter plot can be toggled. Regarding references, a DOI button will appear in blue at the bottom right of each row and clicking it will open the article page.
(3) Alternative Monomers
The number of copolymerization information when one of the two monomers is replaced with another monomer will be displayed in a list with structural formulas and monomer names. The monomers are displayed in order of similarity to the orignal monomer. By clicking on the number of copolymerizations, you can open the detailed page of copolymerization.Search
On the "Copolymers' Search" page, copolymers or copolymerizations can be searched by specifying first and/or second monomer conditions, such as monomer name, molecular formula and number of copolymerizations. Like the monomer search page, this page provides an autocomplete feature (see the monomer search section for more details). A search can be performed by specifying the following conditions.
- Names of first and second monomers: enter a canonical or IUPAC name in text.
- Molecular formulas of first and second monomers: enter a molecular formula in text, regardless of the sequence of atomic symbols. That is, the sequence of pairs of atomic symbols and their numbers is arbitrary; for example, C8H8 and H8C8 are treated as the same molecular formula.
- SMILESs of first and second monomers enter a SMILES in text.
- InChI keys of first and second monomers: enter an InChI key in text.
- Minimum / Maximum number of copolymerizations: enter the minimum and maximum values of the number of copolymerizations as integers.
- Minimum / Maximum r1: enter the minimum and maximum values of first monomer reactivity ratio as integers.
- Minimum / Maximum r2: enter the minimum and maximum values of second monomer reactivity ratio as integers.
Statistics
The "Copolymers' Statistics" page shows 1) basic statistics and 2) histograms of reactivity ratios. As the basic statistics, the number of copolymers, copolymerizations, and DOI references are displayed. As the histograms, the histogram of the number of copolymerizations per copolymer and the histogram of reactivity ratios are displayed.
Data Download
In the near future, we plan to enhance CoPolDB by introducing the capability to download contents. We will offer several methods for data retrieval, including a straightforward CSV file and/or APIs (Application Programming Interfaces). This extension will empower users to leverage CoPolDB data for independent research on their local machines, enabling tasks like constructing machine learning models for predicting reactivity ratios. Furthermore, we envision a future update to CoPolDB that facilitates user contributions by allowing them to register copolymerization results. This enhancement aims to foster collaborative engagement and expand the database's breadth of information.
References
Citation
Contact
CoPolDB was released as a minimum viable service in March 2023. We plan to improve our service based on the opinions of all visitors to this site. If you have questions, comments or suggestions, we are very welcome to hear from you.